- January 28, 2023
- No Comment
- 70 minutes read
Prime editor-mediated correction of a pathogenic mutation in … – Nature.com
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
Advertisement
Carousel with three slides shown at a time. Use the Previous and Next buttons to navigate three slides at a time, or the slide dot buttons at the end to jump three slides at a time.
04 May 2020
Carlos Pinzon-Arteaga, Matthew D. Snyder, … Charles R. Long
10 October 2019
Arisa Ikeda, Wataru Fujii, … Kunihiko Naito
21 October 2019
Andrew V. Anzalone, Peyton B. Randolph, … David R. Liu
18 August 2020
Yuji Yokouchi, Shinichi Suzuki, … Takumi Era
14 October 2020
Haizpea Lasa-Fernandez, Laura Mosqueira-Martín, … Ainara Vallejo-Illarramendi
07 August 2020
Teruaki Tozaki, Aoi Ohnuma, … Shun-ichi Nagata
17 June 2021
Li Xu, Chen Zhang, … Renzhi Han
19 April 2021
Diane J. Sutcliffe, Ashok R. Dinasarapu, … H. A. Jinnah
20 January 2020
Zhiquan Liu, Siyu Chen, … Zhanjun Li
Scientific Reports volume 12, Article number: 12905 (2022)
4195
2
63
Metrics details
An Author Correction to this article was published on 21 September 2022
This article has been updated
Canine hip dysplasia (HD) is a multifactorial disease caused by interactions between genetic and environmental factors. HD, which mainly occurs in medium- to large-sized dogs, is a disease that causes severe pain and requires surgical intervention. However, the procedure is not straight-forward, and the only way to ameliorate the situation is to exclude individual dogs with HD from breeding programs. Recently, prime editing (PE), a novel genome editing tool based on the CRISPR-Cas9 system, has been developed and validated in plants and mice. In this study, we successfully corrected a mutation related to HD in Labrador retriever dogs for the first time. We collected cells from a dog diagnosed with HD, corrected the mutation using PE, and generated mutation-corrected dogs by somatic cell nuclear transfer. The results indicate that PE technology can potentially be used as a platform to correct genetic defects in dogs.
Domestic dogs (Canis lupus familiaris) are the most variable mammalian species1,2. More than 400 breeds have been developed by intense artificial selection from a limited number of founders1,3. Consequently, purebred dogs have a greater risk of suffering from genetic disorders than any other species4. A number of scientific publications have described the health problems of purebred dogs5,6,7,8,9,10,11 and emphasized the need for action9,10,11,12,13,14; the problem has also been highlighted recently in public media15. As a result, many breeders are increasingly using DNA tests to reduce the frequency of deleterious mutations in their breeding programs4. However, no direct treatment has been developed to solve these inherent problems. In particular, canine hip dysplasia (HD) is the most common inherited polygenic orthopedic trait in dog; however, there is still no ideal medical or surgical treatment16.
Genome editing tools, such as CRISPR/Cas9 technology, can be a solution to this genetic problem. In particular, prime editing (PE), a novel and universal precision genome-editing technology has great potential for the correction of pathogenic alleles in purebred dogs. Unlike the conventional CRISPR/Cas9 system, PE does not induce double-strand breaks, which can induce random indel mutations at the target locus. PE was designed to generate a nick in single strands of the target genome locus, and then induce accurate target sequence switching using reverse transcriptase17. While PE was originally developed in human cells17, it has recently been used to develop genome-edited plant varieties and animals, including mice and fruit flies18,19,20. However, there is no report on the use of PE in dogs.
In this study, we used PE to determine whether point mutations causing canine HD can be corrected in canine fibroblasts. Furthermore, we attempted to confirm the possibility of producing genetically modified dogs by somatic cell nuclear transfer (SCNT) using PE-mediated genetically gene-corrected canine fibroblasts.
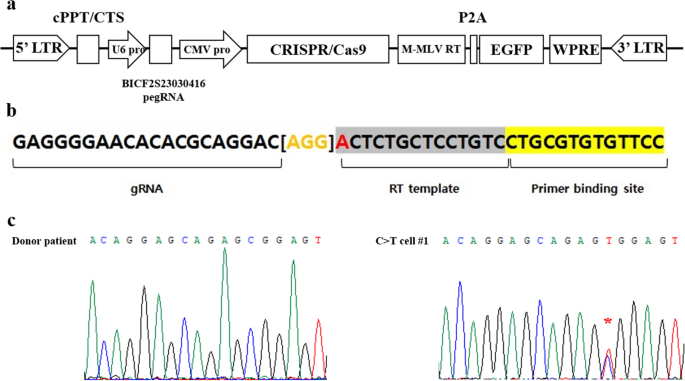
We selected the target point mutation locus for PE-mediated gene correction based on our previous study that identified 25 SNPs correlating with hip dysplasia in dogs21. Among the 25 SNPs, BICF2S23030416 (Supplement Table 1) was selected as the genome editing target in this study because it showed the highest statistical significance for canine HD (p < 0.0001). The BICF2S23030416 SNP is located in an intergenic region on chromosome 4 and is hypothesized to function in regulating MSH homeobox 2 (MSX2). MSX2 has been utilized as a representative marker for cell ossification induction22,23. Our previous report showed that dogs (Labrador retrievers) with HD had a T to C point mutation at the BICF2S23030416 locus among the 25 SNP mutations assessed; therefore, we designed a PE vector to correct the T to C point mutation at this target locus. A lentiviral vector expressing both the PE enzyme (CRISPR/Cas9 nickase fused with M-MLV reverse transcriptase) and a prime editor guide RNA (pegRNA) was constructed and cloned (Fig. 1a). The pegRNA consisted of a primer binding site that could hybridize with sequences near the BICF2S23030416 locus, and a reverse transcriptase template containing the corrected genomic sequence at the point mutation site (Fig. 1b).
Correction of a point mutation in donor cells using prime editor (PE). (a) Schematic of PE vector. It consists of a prime editor that can correct SNP at the BICF2S23030416 locus and EGFP as a reporter. (b) Structure and design of PE guide RNA (pegRNA). The bracketed region in orange color indicates the scaffold for pegRNA17. The nucleotide (A) in red indicates the SNP mutation site. (c) Chromatographic analysis of the donor patient cells and C>T cell #1. The red asterisk indicates the target locus and confirms the C to T sequence correction mediated by PE.
Ear fibroblasts were collected from a Labrador retriever dog diagnosed with HD (donor patient) and cultured in vitro. The T to C point mutation sequence at the BICF2S23030416 locus in the fibroblasts was determined by sequence analysis (Fig. 1c and Supplementary Figure 1, donor patient). Lentiviral particles, which is expressing PE, were transduced into the fibroblasts; the rate of transduction was measured by expression of the enhanced green fluorescent protein (EGFP) reporter gene. After 5 days of culture, gDNA was isolated from the transduced fibroblasts. Sequence analysis confirmed that the PE-treated fibroblasts had a T sequence at the BICF2S23030416 locus, mixed with C, indicating that PE successfully recovered one allele of the point mutation at the target site (Fig. 1c).
After showing that we could correct the point mutation in fibroblasts, we demonstrated that we could produce gene-corrected dogs using SCNT. Among the fibroblasts produced using PE, we selected the ‘C>T cell #1′ for SCNT. In total, 18 reconstructed embryos were generated by SCNT using PE-treated fibroblasts and then surgically transferred into the oviducts of a surrogate mother (Table 1). Pregnancy was detected by ultrasonography at 40 days of gestation, and two puppies weighing 656 g (C>T dog #1) and 585 g (C>T dog #2 were delivered by cesarean section (Fig. 2a). We confirmed the integration of the PE vector by EGFP expression (Fig. 2b) and polymerase chain reaction (PCR) analysis (Fig. 2c). As expected, the C to T gene correction at the BICF2S23030416 locus was confirmed in both puppies (Fig. 2d and Supplementary Figure 1). These results are in line with the sequence analysis data from the PE-treated fibroblasts used as donor cells for SCNT (Fig. 1c). We also performed an in silico analysis of potential off-target loci from C>T dog #1 and C>T dog #2. No off-target mutations were identified in any of the analyzed loci (Table 2).
Generation and verification of gene-corrected dogs using somatic cell nuclear transfer (SCNT). (a) Bright field and (b) enhanced green fluorescent protein (EGFP) expression under UV light of the gene-corrected dogs. (c) PCR analysis of the vector construct integration in donor patient cells and gene-corrected dogs (C>T dog #1 and #2). Only the gene-corrected dogs showed a band at 600 bp. (d) Chromatographic analysis of target sequences from the donor patient (cells) and gene-corrected dogs. Blue star asterisks indicate SNP of donor patient and red star asterisks indicate corrected SNP sequence in C>T dogs #1 and #2.
In the present study, we successfully generated two gene-corrected dogs, cloned from a dog diagnosed with HD, using PE technology for the first time. HD is a musculoskeletal disorder caused by an unstable connection between the femoral head and acetabulum and is accompanied by severe pain. It is a common disorder in medium- to large-sized dogs, and is known to cause osteoarthritis, lameness and decreased mobility24. HD is a polymorphic disease caused by a combination of genetic and environmental factors25. Thus, to reduce the prevalence of HD, breeding strategies incorporating screening schemes are widely used24. However, studies that eliminate the cause of HD by directly controlling the causative gene have not yet been reported in dogs.
PE technology is effective and a potential solution for correcting genetic mutations in specific canine breeds. It is a simple and highly efficient gene correction system compared to that of CRISPR/Cas9-mediated homology directed repair (CRISPR-HDR)17,18,19. The CRISPR-HDR method is dependent on cell division events and requires an additional donor DNA template to correct genetic mutations. PE overcomes the shortcomings of CRISPR-HDR; it can be performed at any stage of the cell cycle and does not require additional donor DNA17. Therefore, PE is expected to be a very useful tool, enabling precise target sequence correction at specific loci in dogs. In addition, we also analyzed sequences from the potential off-target loci and did not find any unexpected mutations. The off-target analysis results revealed that the PE system is specific in canine cells. These findings are in line with previous studies demonstrating that the PE-mediated base conversion is highly specific17,18.
We corrected a single SNP from a dog with the HD phenotype. However, due to multiple SNP mutations are contributed to the HD, additional gene correction at the other SNP loci related to HD might be needed to generate a fully HD-recovered canine breed. We regarded the current study is the starting point to overcome HD of purebred dog. Thus, we integrated our PE vector into the genome of our gene corrected dogs and plan to perform further studies focused on correcting the additional SNPs. Integrated PE system will induce spontaneous nickase activity at the target site. In the current study, we did not find any indel mutation from our sequencing results; however, a more stable form of PE, such as RNP, can be recommended in further studies. Precise editing of pathogenic SNP in dog also provides valuable information for understanding the role of each SNP as it relates to HD. Since canine HD is remarkably similar in clinical expression and pathogenesis to that of human HD26, information gleaned from gene-corrected dogs may be very useful for understanding human HD. Thus, PE may be a very useful tool for generating genome-edited dog models to study human diseases.
In conclusion, we successfully confirmed the feasibility of PE in dogs and produced HD-related gene-corrected dogs using PE. To the best of our knowledge, this is the first study to adapt PE for use in a canine system. Further studies to analyze gait, behavior, and mobility of the current gene-corrected dogs, and the generation of additional gene-corrected dogs, are needed to understand the relationship between each SNP and HD.
The experimental procedures and methods used in this study were approved by the Animal Welfare and Ethics Office (2019012A-CNU-174), Chungnam National University, Daejeon, and performed according to “The Guide for the Care and Use of Laboratory Animals” published by IACUC of Chungnam National University. Female mixed dogs from 2 to 6 years of age were used in this study as oocyte donors and embryo transfer recipients. The dogs were housed indoors and fed once daily with water ad libitum. All methods are reported in accordance with ARRIVE guidelines (https://arriveguidelines.org) for the reporting of animal experiments in the Methods section.
The vector for PE was purchased from Addgene (Watertown, MA, USA: #135955) and modified to correct HD-related SNPs. Briefly, the CMV promoter was obtained by PCR using the primer sets 5′-gaattcttgacattgattattgactag-3′ and 5′-tctagaaatttcgataagccagtaagc-3′, and inserted into the vector by EcoRI and XbaI (NEB Inc., MA, USA: #R0101M and #R0145M) enzyme cuts. The pegRNA targeting the HD locus was newly synthesized and then added to the vector using PacI (NEB Inc., MA, USA: #R0547S) and EcoRI. Finally, the vector was confirmed through sequencing. The lentiviral particles of PE vector were produced by commercial vendor (Lugen SCI, Inc., Bucheon, South Korea).
Fibroblasts were collected from the ears of an 18-month-old Labrador retriever diagnosed with HD (donor patient). The primary fibroblasts were cultured in vitro using culture medium composed of DMEM-GlutaMAX, 15% fetal bovine serum, and 1% penicillin/streptomycin solution (GIBCO, Inc.). For transduction, 100 multiplicity of infection (MOI) of the PE lentiviral particles, containing 1 μg/mL of polybrene, was transduced into 1 × 105 fibroblasts per a well of 12-well plate. Transgene expression was confirmed by EGFP and integration of the vector was confirmed by sequence analysis.
We collected mature oocytes from dogs as described previously27. The concentration of progesterone in the blood was measured to optimize the hormone concentration for harvesting mature oocytes. After confirming the time of estrus, blood was collected, and progesterone was measured using VET Chroma (ANIVET Inc., Chuncheon, South Korea). When the analyzed progesterone level was in the range of 4–7 ng/mL we considered that day as ovulation. Three days after ovulation, mature oocytes were surgically collected. During the procedure, all dogs were treated with ketamine and xylazine at a concentration of 6 mg/Kg, and anesthesia was maintained with 2% isoflurane. After exposing the ovary and uterus, a 24G intravenous catheter was inserted into the oviductal lumen near the uterotubal junction, and the culture medium was flowed to collect mature oocytes. The culture medium was prepared by adding 2 mM NaHCO3, 1% penicillin/streptomycin, 0.5% bovine serum albumin, and 10% FBS to medium 199 containing 25 mM HEPES.
For generating gene corrected dogs, SCNT followed by embryo transfer was performed following the method described elsewhere27. Briefly, in vivo matured oocytes with the first polar body were used for micromanipulation. Metaphase chromosomes were removed by aspiration from the oocytes. A single cell (C>T cell) was transferred into the perivitelline space of an enucleated oocyte, and each donor cell-cytoplast couplets were fused by two pulses of direct current (24–26 V for 15 μsec) using an Electro-Cell fusion apparatus. The fused SCNT embryos were chemical activated by incubating with 10 μM calcium ionophore (Sigma) and then 1.9 mM 6-dimethylaminopurine (6-DMAP). The activated SCNT embryos were surgically transferred into the oviducts of estrus-synchronized surrogates. Pregnancy was confirmed by ultrasonography at 30 days after embryo transfer.
Transgene integration into the genome of transduced fibroblasts and gene-corrected dogs was confirmed by PCR. The PCR primers used to validate the Cas9 sequence in the vector were 5′-catcgctattaccatggtgat-3′ and 5′-ctcttgcagatagcagatcc-3′. These primer sets detected the linkage between the CMV promoter and dCas9 of the vector used in this study. Sequencing of the target locus was performed to validate the PE-mediated gene correction. The sequencing primers used were 5′-gacgccaagggagcagatatt-3′ and 5′-cctctcttatgagaacagcat-3′ (Bioneer Inc., Daejeon, South Korea). In addition, TA cloning was performed for accurate sequencing analysis using the products generated through PCR (Supplement Fig. 1). PCR products and T vector (Promega Inc., WI, USA: #A1360) were mixed at a ratio of 1:3, and DNA was isolated and purified from the bacteria colony generated by ligation (NEB Inc., MA, USA: #M0202) to extract DNA. After confirming the extracted DNA with EcoRI restriction enzyme, sequencing analysis was performed.
Potential off-target loci were determined in silico using Cas-OFFinder (http://www.rgenome.net/cas-offinder/). We selected two potential off-target loci with two mismatches and another three loci with three mismatches compared to the genomic target sequences of pegRNA used in the study. The potential off-target loci were PCR amplified with genomic DNA from the C>T dog #1 and C>T dog #2 and sequencing analysis was performed (Supplementary Table 2).
In conducting this study, we collected cells from a retriever with hip dysplasia, and this was done after explaining the study to the retriever owner and consent was obtained. The experimental procedures and methods used in this study were approved by the Animal Welfare and Ethics Office (CNU-01090) of Chungnam National University, Daejeon, and performed according to the Guide for the Care and Use of Laboratory Animals published by the IACUC of Chungnam National University. All methods are reported in accordance with ARRIVE guidelines (https://arriveguidelines.org) for the reporting of animal experiments in the Methods section.
Not applicable.
The datasets generated and/or analysed during the current study are not publicly available due to some data required for our further studies but are available from the corresponding author on reasonable request.
A Correction to this paper has been published: https://doi.org/10.1038/s41598-022-20187-7
Ostrander, E. A. et al. Dog10K: An international sequencing effort to advance studies of canine domestication, phenotypes and health. Natl. Sci. Rev. 6, 810–824 (2019).
Article CAS Google Scholar
Vilà, C., Maldonado, J. E. & Wayne, R. K. Phylogenetic relationships, evolution, and genetic diversity of the domestic dog. J. Hered. 90, 71–77 (1999).
Article Google Scholar
Plassais, J. et al. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 10, 1–14 (2019).
Article CAS Google Scholar
Mellersh, C. DNA testing and domestic dogs. Mamm. Genome 23, 109–123 (2012).
Article Google Scholar
Oberbauer, A., Belanger, J., Bellumori, T., Bannasch, D. & Famula, T. Ten inherited disorders in purebred dogs by functional breed groupings. Canine Genet. Epidemiol. 2, 1–12 (2015).
Article Google Scholar
Nicholas, F. W., Wade, C. M. & Williamson, P. Disorders in pedigree dogs: assembling the evidence. Vet. J. 183, 8–9 (2009).
Article Google Scholar
Nicholas, F. W., Crook, A. & Sargan, D. R. Internet resources cataloguing inherited disorders in dogs. Vet. J. 189, 132–135 (2011).
Article Google Scholar
Ginja, M., Silvestre, A., Gonzalo-Orden, J. & Ferreira, A. Diagnosis, genetic control and preventive management of canine hip dysplasia: A review. Vet. J. 184, 269–276 (2010).
Article CAS Google Scholar
Hedhammar, Å. European strategies to enhance canine genetic health. Eur. J. Companion Anim. Pract. 9, 93 (1999).
Google Scholar
Hedhammar, Å. Actions by FCI and WSAVA to promote canine genetic health. Eur. J. Companion Anim. Practice 15, 22–25 (2005).
Google Scholar
McGreevy, P. D. & Nicholas, F. Some practical solutions to welfare problems in dog breeding. ANIMAL WELFARE-POTTERS BAR- 8, 329–342 (1999).
Google Scholar
Indrebø, A. Breeding healthy dogs–A breeder’s perspective. Eur. J. Companion Anim. Practice 15, 17–21 (2005).
Google Scholar
Asher, L., Diesel, G., Summers, J. F., McGreevy, P. D. & Collins, L. M. Inherited defects in pedigree dogs. Part 1: Disorders related to breed standards. Vet. J. 182, 402–411 (2009).
Article Google Scholar
Summers, J. F., Diesel, G., Asher, L., McGreevy, P. D. & Collins, L. M. Inherited defects in pedigree dogs. Part 2: Disorders that are not related to breed standards. Vet. J. 183, 39–45 (2010).
Article Google Scholar
Hedhammar, Å. A., Malm, S. & Bonnett, B. International and collaborative strategies to enhance genetic health in purebred dogs. Vet. J. 189, 189–196 (2011).
Article Google Scholar
Ginja, M., Gaspar, A. R. & Ginja, C. Emerging insights into the genetic basis of canine hip dysplasia. Vet. Med. Res. Rep. 6, 193 (2015).
Google Scholar
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Article ADS CAS Google Scholar
Liu, Y. et al. Efficient generation of mouse models with the prime editing system. Cell Discov. 6, 1–4 (2020).
Article Google Scholar
Jiang, Y.-Y. et al. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 21, 1–10 (2020).
Article Google Scholar
Bosch, J. A., Birchak, G. & Perrimon, N. Precise genome engineering in Drosophila using prime editing. Proc. Natl. Acad. Sci. 118, e2021996118 (2021).
Article CAS Google Scholar
Choi, B. H., Kim, T. H., Lee, S. H. & Im, S. K. SNP for diagnosing hip dysplasia in dog and uses thereof. Republic of Korea patent 10-2012-0043793 (2013).
Ichida, F. et al. Reciprocal roles of MSX2 in regulation of osteoblast and adipocyte differentiation. J. Biol. Chem. 279, 34015–34022 (2004).
Article CAS Google Scholar
Matsubara, T. et al. BMP2 Regulates Osterix through Msx2 and Runx2 during Osteoblast Differentiation∗. J. Biol. Chem. 283, 29119–29125 (2008).
Article CAS Google Scholar
Lewis, T. W., Blott, S. C. & Woolliams, J. A. Genetic evaluation of hip score in UK Labrador Retrievers. PLoS ONE 5, e12797 (2010).
Article ADS Google Scholar
Wang, S. et al. Genetic correlations of hip dysplasia scores for Golden retrievers and Labrador retrievers in France, Sweden and the UK. Vet. J. 226, 51–56 (2017).
Article ADS CAS Google Scholar
Zhou, Z. et al. Differential genetic regulation of canine hip dysplasia and osteoarthritis. PLoS ONE 5, e13219 (2010).
Article ADS Google Scholar
Lee, J. H. et al. Effect of acteoside as a cell protector to produce a cloned dog. PLoS ONE 11, e0159330 (2016).
Article Google Scholar
Download references
We thank the Dr. Bong-Hwan Choi of the National Institute of Animal Sciences for allowing us to use the results of Hip dysplasia. We would also like to thank the owner for consenting to the study of a retriever with Hip Dysplasia in conducting this study and for helping to collect the cells.
This work was carried out with the support of “MK biotech” and “Cooperative Research Program of Center for Companion Animal Research (Project No. PJ01398702)” Rural Development Administration, Republic of Korea.
Laboratory of Animal Reproduction and Physiology, Division of Animal and Dairy Science, College of Agriculture and Life Science, Chungnam National University, Daejeon, 34134, Korea
Dong Ern Kim, Ji Hye Lee, Kuk Bin Ji, Eun Ji Lee, Chuang Li, Hyun Ju Oh & Min Kyu Kim
MK Biotech Inc., Daejeon, 34134, Korea
Kang Sun Park & Min Kyu Kim
National Institute of Animal Science, Wanju, 55365, Korea
Seung Hoon Lee
ToolGen Inc., Seoul, 08501, Korea
Okjae Koo
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
D.E.K., O.J.K. and M.K.K. conceived and designed the study. D.E.K., J.H.L., K.B.J., E.J.L., C.L. and K.S.P. was responsible for performed the experiment. D.E.K., S.H.L., O.J.K. and M.K.K. analyzed the data. D.E.K., O.J.K. and M.K.K. wrote the paper. H.J.O., O.J.K. and M.K.K. edited and reviewed the manuscript. All authors read and approved the manuscript.
Correspondence to Okjae Koo or Min Kyu Kim.
The authors declare no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in Figure 1. As a result of an error during figure assembly, the C>T cell #1 panel in Figure 1c was a duplication of C>T dog #1 panel in Figure 2d. Figure 1 was replaced to show the correct data.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
Kim, D.E., Lee, J.H., Ji, K.B. et al. Prime editor-mediated correction of a pathogenic mutation in purebred dogs. Sci Rep 12, 12905 (2022). https://doi.org/10.1038/s41598-022-17200-4
Download citation
Received: 23 November 2021
Accepted: 21 July 2022
Published: 28 July 2022
DOI: https://doi.org/10.1038/s41598-022-17200-4
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Nature Reviews Genetics (2022)
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Advertisement
© 2023 Springer Nature Limited
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

